Bio & Health Informatics Group
The Bio & Health Informatics Group is developing new methods and systems for biological and health data analysis, modelling, prediction and understanding.
Current projects
The lab is working on several projects, including:
SNN techniques are used to model and analyse static and dynamic transcriptomic and metabolomics data by means of knowledge extraction from a gene interaction network.
The Brain-Like Artificial Intelligence (BLAI) is pioneered by Professor Nikola Kasabov and here it is applied to a specific application.
This SNN system propose novel research which is current under development. This novel system for bioinformatics data analysis is being applied to:
- Static and Dynamic Transcriptomics Data Modelling and Analysis
- Metabolomics Data Modelling and Analysis
- Nutrigenomics Data Modelling and Analysis
- Feature Selection Methods for Gene Expression Data Analysis
This SNN system can also be used to understand the biochemical mechanims that govern synaptic plasticity in the human brain and regulate cognitive by mean of glutamatergic and GABAergic bidirectional activity.
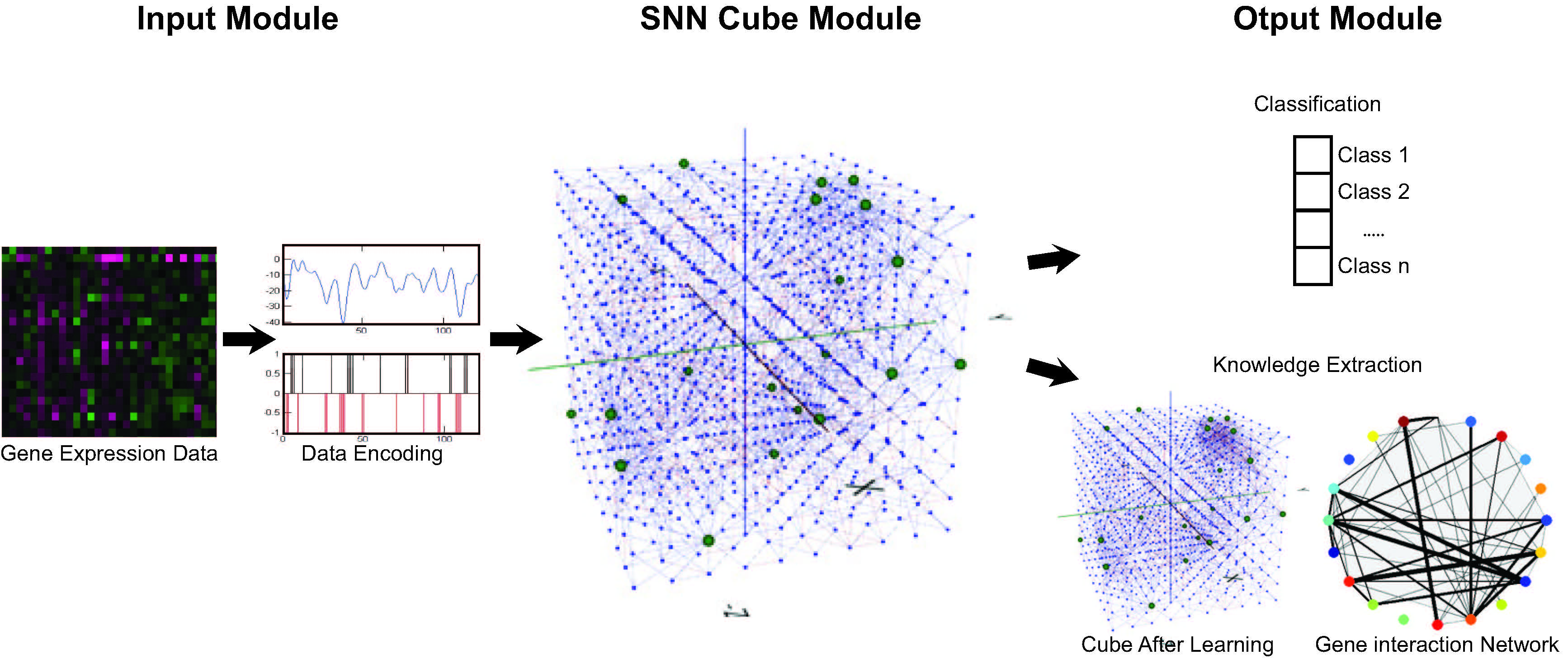
Schematic representation of the SNN system for bioinformatics data modelling and analysis.
Related papers, patents and benchmarking
The proposed methods and systems, when compared with traditional statistical and machine learning methods, showed superior results in the following aspects:
- Better data analysis and classification accuracy;
- Better visualisation of the created models, with a possible use of VR;
- Better understanding of the data and the processes that are measured through a gene interaction network;
- Enabling new information and knowledge discovery through meaningful interpretation of the models and the genetics process that regulate the data.
See also some of the related papers:
- Kasabov, N., Scott, N. M., Tu, E., Marks, S., Sengupta, N., Capecci, E., Othman, M., Gholoami Doborjeh, M., Murli, N., Hartono, R., Espinosa-Ramos, J. I., Zhou, L., Alvi, F., Wang, G., Taylor, D., Feigin, V., Gulyaev, S., Mahmoud, M., Hou, Z. G., Yang, J. (2016). Evolving spatio-temporal data machines based on the NeuCube neuromorphic framework: design methodology and selected applications. Neural Networks, 78, 1-14
- Espinosa-Ramos, J.I., Capecci, E., Kasabov, N. (2017). A Computational Model of Neuroreceptor Dependent Plasticity (NRDP) Based on Spiking Neural Networks. IEEE Transactions on Cognitive and Developmental Systems. Special Issue on Spiking Neural Networks. Available online 22/11/2017. DOI:10.1109/TCDS.2017.2776863
- Koefoed, L., Capecci, E. and Kasabov, N. (2018, July). Time Series Analysis of rVSV-ZEBOV trial data using TMRMR-C and NeuCube. 2018 International Joint Conference on Neural Networks (IJCNN). Rio De Janeiro, Brazil, 8-13 July (accepted). IEEE
R&D system
For this project, an R&D system has been developed based on NeuCube. The system can be obtained subject to licensing agreement.
Developer
SNN-based Personalised Modelling is an advanced data analytic that offers improved personal outcome prediction, personalisation of treatment and understanding through identifying the most predictive factors for a person.
The Brain-Like Artificial Intelligence (BLAI) is pioneered by Professor Nikola Kasabov and here it is applied to a specific application.
NeuCube-based personalised modelling is an advanced data modelling system that offers improved personal outcome prediction, personalisation of treatment and understanding through identifying the most predictive factors for a person across various types of data, including both static and dynamic (temporal) types.
This system can be applied to brain data, environmental and ecological data, social and financial data etc.
The proposed NeuCube personalised modelling is performed based on the following diagram:
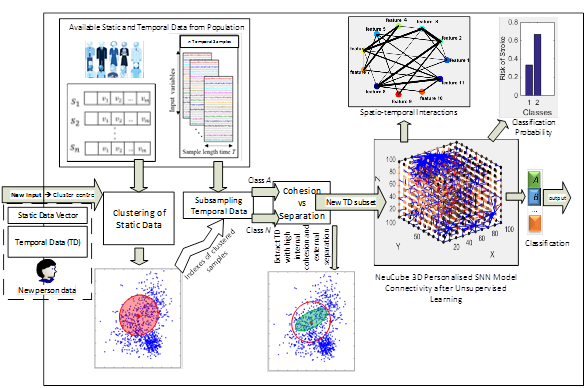
The NeuCube personalised modelling for integrated static and dynamic data.
Related papers, patents and benchmarking
The proposed methods and systems, when compared with traditional statistical and machine learning methods, showed superior results in the following aspects:
- Better data analysis and classification/regression accuracy (by 10 to 40%);
- Better visualisation of the created models, with a possible use of VR;
- Better understanding of the data and the processes that are measured;
- Enabling new information and knowledge discovery through meaningful interpretation of the models.
See also some of the related papers:
- Doborjeh, M. G., & Kasabov, N. (2016, July). Personalised modelling on integrated clinical and EEG spatio-temporal brain data in the NeuCube spiking neural network system. In Neural Networks (IJCNN), 2016 International Joint Conference on (pp. 1373-1378). IEEE.
- Kasabov, N. K., Hou, Z. G., Feigin, V., & Chen, Y. (2014). U.S. Patent Application No. 14/914,326.
- N.Kasabov, V.Feigin, Z.Hou, Y.Chen, Improved method and system for predicting outcomes based on spatio/spectro-temporal data, PCT patent WO2015/030606 A2, US2016/0210552 A1. Granted/Publication date: 21 July 2016.
- N.Kasabov, Data Analysis and Predictive Systems and Related Methodologies, US patent 9,002,682 B2, 7 April 2015.
R&D system
For this project, an R&D system has been developed based on NeuCube. The system can be obtained subject to licensing agreement.
Developer
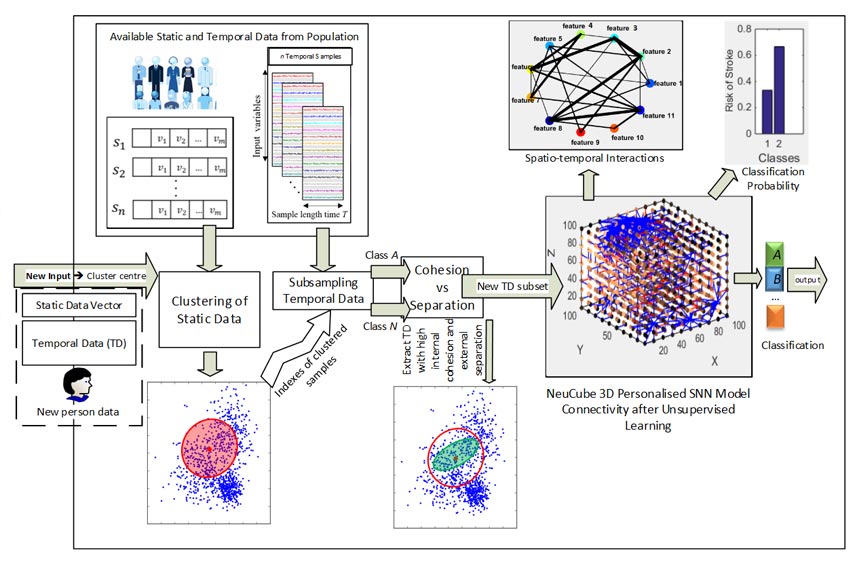
SNN-based personalised modelling for prediction of risk of stroke using static vector-based data and dynamic environmental time series.
The Brain-Like Artificial Intelligence (BLAI) is pioneered by Professor Nikola Kasabov and here it is applied to a specific application.
This project develops a novel personalised modelling method and system aimed at developing computational prognostic or diagnostic systems for risk of stroke. The proposed method is based on integration of different data processing techniques for appropriate selection of features (variables) and neighbouring samples. This has the potential to identify the most important characteristics of an individual through personalised profiling and improves the classification/prediction of output (risk of stroke event) as compared to global modelling. In this section, instead of building a global model and training it with the whole population in data, for every person we will build a personalised SNN model (PSNN) to train it only on a subset of data which belongs to individuals who have similar integrated static clinical factors and dynamic environmental data.
The proposed NeuCube personalised modelling is performed based on the following steps:
- For a new person data xi
- A cluster of K number of patients (had stroke) who have small distance to xi will be selected (according to static data of stroke). Clustering of integrated static-dynamic data is performed using the new algorithm dWWKNN (dynamic weighted–weighted distance K-nearest neighbours). For a new individual xi, we rank the contribution of each of the k neighbouring samples based on integrated static-dynamic distance to the xi, giving greater rank to closer neighbours.
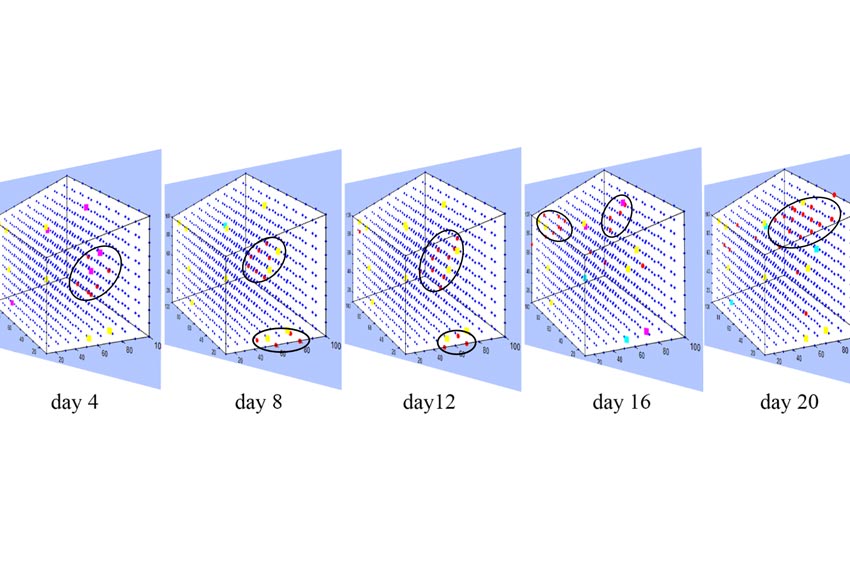
- For every member in this cluster, the date of stork will be found and two environmental samples will be extracted with respect to the below time intervals:
- High risk of stroke: environmental data of 20 days (a parameter that can be adjusted by the user) before the stroke event
- Low risk of stroke: environmental data of 20 days in the interval of 60 to 40 days before the stroke event
- K*2 environmental samples will be used for creating a personalised SNN model, training and testing.
- The output prediction provides a probability whether the person xi is in high risk of stroke or low risk.
Related papers, patents and benchmarking
The proposed methods and systems, when compared with traditional statistical and machine learning methods, showed superior results in the following aspects:
- Identification of the most important characteristics of an individual through personalised profiling;
- Improved classification/prediction of output (risk of stroke event) as compared to global modelling;
- Better visualisation of the created models, with a possible use of VR;
- Better understanding of the data and the processes that are measured;
- Enabling new information and knowledge discovery through meaningful interpretation of the models.
See also some of the related papers:
- Kasabov, N. K. (2014). NeuCube: A spiking neural network architecture for mapping, learning and understanding of spatio-temporal brain data. Neural Networks, 52, 62-76.
- Kasabov, N., Feigin, V., Hou, Z. G., Chen, Y., Liang, L., Krishnamurthi, R., Othman, M., Parmar, P. (2014). Evolving spiking neural networks for personalised modelling, classification and prediction of spatio-temporal patterns with a case study on stroke. Neurocomputing, 134, 269-279. doi:10.1016/j.neucom.2013.09.049
- Kasabov, N., Feigin, V., Hou, Z. -G., Chen, Y., Liang, L., Krishnamurthi, R., Parmar, P. (2014). Evolving spiking neural networks for personalised modelling, classification and prediction of spatio-temporal patterns with a case study on stroke. Neurocomputing, 134, 269-279.
- Feigin, V., P.Parmar, S. Barker-Collo, D.A Bennett, C.S Anderson, A. G Thrift, B. Stegmayr, P. M Rothwell, M.Giroud, Y. Bejot, P. Carvil, R.Krishnamurthi, N.Kasabov (2014). Geomagnetic Storms Can Trigger Stroke. Stroke, STROKEAHA-113.
- N.Kasabov, V.Feigin, Z.Hou, Y.Chen, Improved method and system for predicting outcomes based on spatio/spectro-temporal data, PCT patent WO2015/030606 A2, US2016/0210552 A1. Granted/Publication date: 21 July 2016.
- N.Kasabov, Data Analysis and Predictive Systems and Related Methodologies, US patent 9,002,682 B2, 7 April 201
R&D system
For this project, an R&D system has been developed based on NeuCube. The system can be obtained subject to licensing agreement.
Developer
Personalised modelling is concerned with the creation of Individual model from the available data to help estimating an unknown outcome for an individual.
The Brain-Like Artificial Intelligence (BLAI) is pioneered by Professor Nikola Kasabov and here it is applied to a specific application.
This project develops methods and systems for personalised modelling (PM). Personalised modelling aims to create a unique computational diagnostic model for each individual, taking into considering the fact that each individual is different and the most effective treatment for an individual can be achieved by a detailed analysis of the data available for that particular individual. The rationale behind personalised modelling is that each person is different and hence they have different requirements and responses to the same treatment. The most effective treatment for an individual can thus be achieved only by the detailed analysis of data available for the patient.
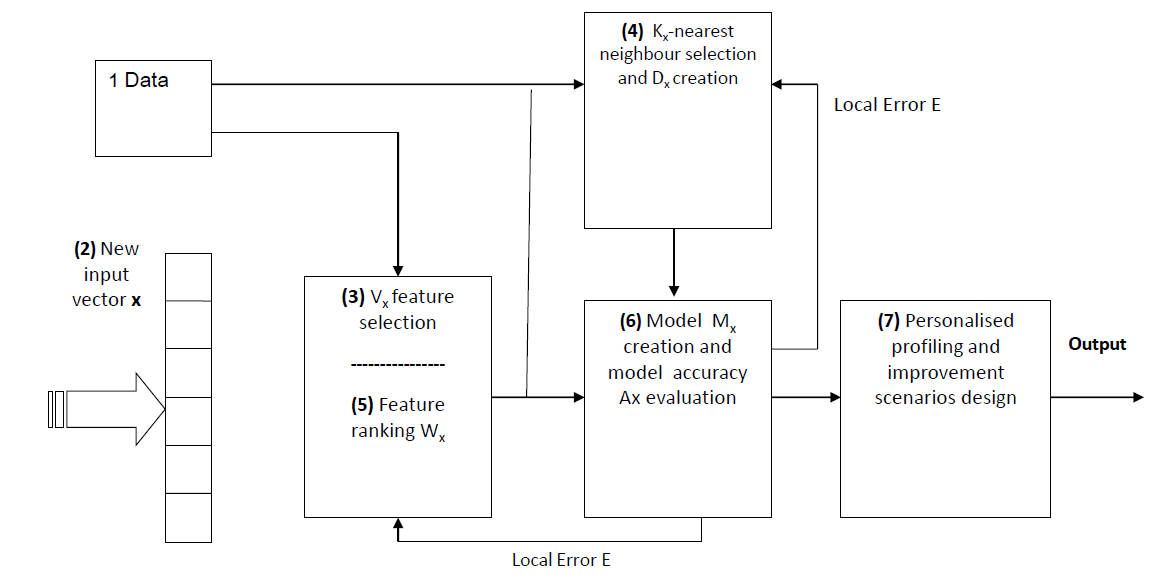
Conceptual Framework for Personalised Modelling for Medical Decision Support.
Related papers, patents and benchmarking
The proposed methods and systems, when compared with traditional statistical and machine learning methods, showed superior results in the following aspects:
- Better data analysis and classification accuracy (~10%);
- Better prognostic accuracy and a computed personalised profile;
- It enables better visualisation of the risk factors for a particular sample.
See also some of the related papers:
- Kasabov, N., Feigin, V., Hou, Z., Chen, Y. (2016). Improved method and system for predicting outcomes based on spatio/spectro-temporal data, PCT patent WO2015/030606 A2, US2016/0210552 A1. Granted/Publication date: 21 July 2016.
- Kasabov, N. (2015). Data Analysis and Predictive Systems and Related Methodologies, US patent 9,002,682 B2, 7 April 2015.
- Kasabov, N. (2007). Global, local and personalised modeling and pattern discovery in bioinformatics: An integrated approach. Pattern Recognition Letters, 28(6), 673-685.
- Kasabov, N., & Hu, Y. (2010). Integrated optimisation method for personalised modelling and case studies for medical decision support. International Journal of Functional Informatics and Personalised Medicine, 3(3), 236-256.
R&D system
The software architecture mentioned above is being implemented in Python language as Open Source Software which is currently under development and will be available subject to licensing in 2018.
Developer
Our research groups
The research and development work done by KEDRI's founding director, Professor Nikola Kasabov, and his team is organised into six areas of research.