Brain Data Analysis Group
The Brain Data Analysis Group was established in November 2015. Its main objectives are to offer contemporary tools and systems for brain data modelling, analysis and understanding, including EEG data, fMRI data, integrated static and dynamic brain data.
Its applications span across neuromarketing, EEG data modelling, fMRI data modelling and brain computer interfaces (BCI).
Current projects
The lab is working on several projects, including:
This R&D system uses spiking neural networks for the analysis of Neuromarketing data.
The Brain-Like Artificial Intelligence (BLAI) is pioneered by Professor Nikola Kasabov and here it is applied to a specific application.
In neuromarketing two projects have been conducted that result in two R&D systems developed and available subject to licensing:
- Study of peri-perceptual patterns with spiking neural networks
- Attentional Bias Recognition
Related papers and benchmarking
The proposed methods and systems, when compared with traditional statistical and machine learning methods, showed superior results in the following aspects:
- Better data analysis and classification/regression accuracy (by 10 to 40%);
- Better visualisation of the created models, with a possible use of VR;
- Better understanding of the data and the processes that are measured;
- Enabling new information and knowledge discovery through meaningful interpretation of the models.
See also some of the related papers:
- Kasabov, N. K. (2014). NeuCube: A spiking neural network architecture for mapping, learning and understanding of spatio-temporal brain data. Neural Networks, 52, 62-76.
- ZG Doborjeh, N Kasabov, MG Doborjeh Attentional Bias Pattern Recognition in Neuromarketing using EEG Data and the NeuCube Evolving Spatio-Temporal Data Machine Based on Spiking Neural Networks", Cognitive Computation, 2017.
R&D system
For the Neuromarketing projects, two R&D systems have been developed based on NeuCube. The systems can be obtained for R&D subject to licensing agreement.
Neuromarketing data
Download our neuromarketing data
Developer
This methodology uses spiking neural networks for the classification, analysis and understanding of spatio-temporal brain EEG data. Some of the applications are: predicting the outcome of a drug treatment, neurological event diagnose and neurorehabilitation.
The Brain-Like Artificial Intelligence (BLAI) is pioneered by Professor Nikola Kasabov and here it is applied to a specific application.
Some of the applications of this system have been developed thorough a collaboration between KEDRI and other international and national institutes such as the Faculty of Health and Environmental Sciences of AUT and the Neulab of the Mediterranean University of Reggio Calabria.
EEG records brain signals through electrodes on the scalp and is widely used in research, as it is non-invasive and has good temporal and spatial resolution. However, EEG systems have been criticized, because of the time consuming and complex training period for the potential user. Nevertheless, EEG is the most commonly collected data for the study of brain processes.
Recently, a growing number of tools have been developed to process this data. Classification of EEG signals is often done with standard statistical and artificial intelligence methods. All these techniques often require extensive preprocessing of the data and also signal averaging. More efficient techniques are required, as the EEG can provide powerful information that, if understood, can be used to stage and diagnose cognitive impairments, predict neurological disorders and dementia, help developing device that can support the neurorehabilitation process, etc.
In this respect, a major contribution to research can be brought about by a much more efficient methodology based on machine learning techniques that can be used to evaluate the EEG signals of a person with a deeper understanding of the underlying brain processes.
Here, we have developed a new spiking neural network methodology and a system for the classification and analysis of EEG data. This system can be used to model, recognise and understand complex EEG data recorded during different scenarios. The approach used in this research is based on the spiking neural networks NeuCube architecture.
The NeuCube-based approach leads to faster data processing, improved accuracy of the EEG data classification and improved understanding of the brain information and the cognitive processes that generated it.
All of this proves that the new proposed methodology and system can constitute an important contribution to the IS field of research, as it allows for many applications to be developed, such as: new types of BCI; early detection of cognitive decline to be used by clinicians in everyday diagnosis; neuromarketing based on EEG data; pain detection; cognitive games; and many more.
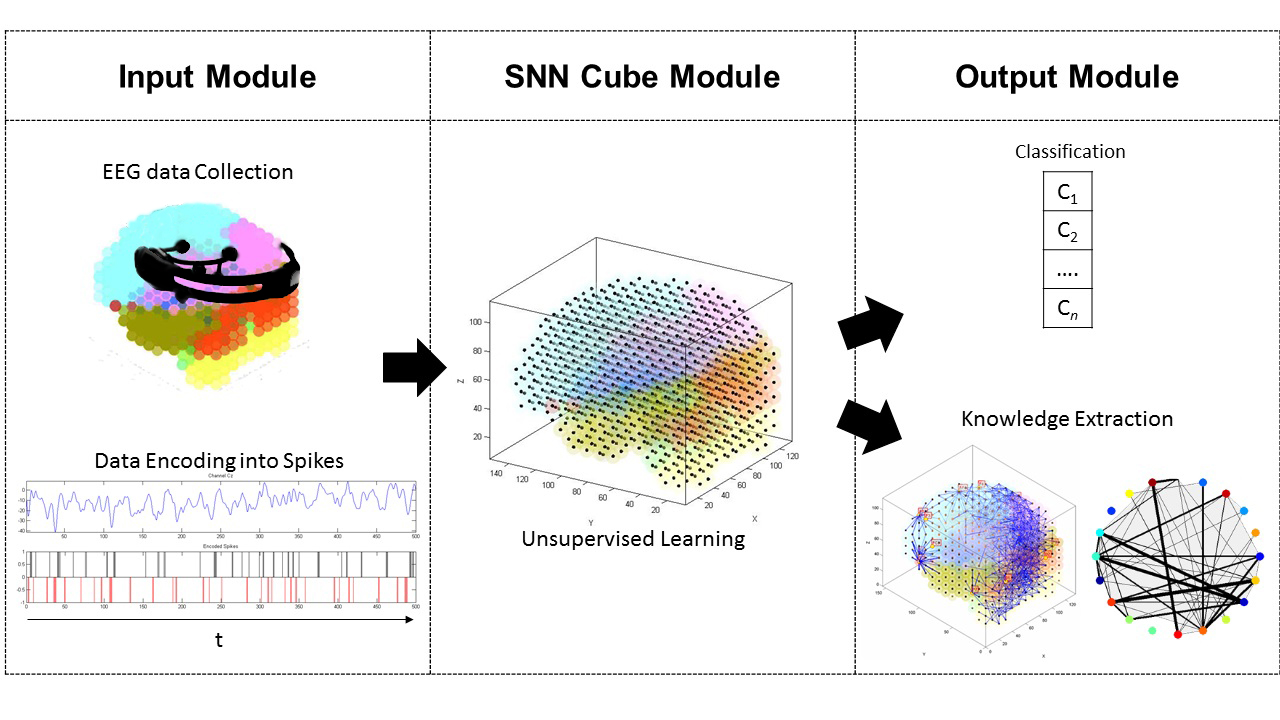
The image shows the NeuCube architecture for EEG data classification and knowledge extraction. The picture shows the NeuCube three principal sub-modules: the input encoding sub-module, where EEG data is encoded into trains of spikes that are then presented to the main sub-module, the SNN cube; the NeuCube main sub-module, where time and space characteristics of the data are captured and learned; and the output sub-module for data classification and new knowledge discovery from the SNNc visualization. Image from (Kasabov, 2014a).
Related papers and benchmarking
The proposed methods and systems, when compared with traditional statistical and machine learning methods, as demonstrated to offer several avantages, such as:
- No need of preprocessing of the data (such as normalization, scaling, smoothing, etc.);
- Fast learning of EEG data with one iteration data propagation only;
- Better classification accuracy (see table below from Kasabov & Capecci, 2014);
- Ability to adapt to new data and new classes through incremental learning;
- Connectivity analysis of the model after unsupervised training of EEG data from different groups of subjects/classes to extract new knowledge and to study the brain regions involved;
- Connectivity analysis for monitoring the treatment of patients and the state of their cognitive brain activity;
- Classification and connectivity analysis for predicting patient response to treatment;
- The model capability for use in brain-like mapping, to study the specific characteristics of neurological events, revealing the area of the brain where it occurs. Leading eventually to future prediction of neurological events based on the initial information;
- The model capability for use in neuromorphic learning, for diagnostic decision support systems, personalised prognosis, and early prediction of a neurological event;
Measure | ELM | NeuCube | SVM | NeuCube |
|---|---|---|---|---|
Accuracy (%) | 87% | 92% | 78% | 84% |
Liang, N. Y., Saratchandran, P., Huang, G. B., & Sundararajan, N. (2006). Classification of mental tasks from EEG signals using extreme learning machine. International journal of neural systems, 16(01), 29-38.
See also some of the related papers:
- Kasabov, N., & Capecci, E. (2015). Spiking neural network methodology for modelling, classification and understanding of EEG spatio-temporal data measuring cognitive processes. Information Sciences, 294, 565-575.
- Capecci, E., Wang, G., Kasabov, N. (2015). Analysis of connectivity in a NeuCube spiking neural network trained on EEG data for the understanding and prediction of functional changes in the brain: A case study on opiate dependence treatment, Neural Networks, vol.68, 62-77. doi:10.1016/j.neunet.2015.03.009
- Doborjeh, M. G., Wang, G. Y., Kasabov, N. K., Kydd, R., & Russell, B. (2016). A spiking neural network methodology and system for learning and comparative analysis of EEG data from healthy versus addiction treated versus addiction not treated subjects. IEEE Transactions on Biomedical Engineering, 63(9), 1830-1841.
- Taylor, D., Scott, N., Kasabov, N., Capecci, E., Tu, E., Saywell, N.,Chen, Y., Hu, J., Hou, Z. G. (2014, July). Feasibility of neucube snn architecture for detecting motor execution and motor intention for use in bci applications. In Neural Networks (IJCNN), 2014 International Joint Conference on (pp. 3221-3225). IEEE.
- Capecci, E., Espinosa-Ramos, J.I., Mammone, N., Kasabov, N., Duun-Henriksen, J., Kjaer, T. W., Campolo, M., La Foresta, F., Morabito, F.C. (2015). Absence Epilepsy Seizure Data in the NeuCube Evolving Spiking Neural Network Architecture. International Joint Conference on Neural Networks (IJCNN). Killarney, Ireland, 12-17 July. IEEE.
- Capecci, E., Doborjeh, Z. G., Mammone, N., La Foresta, F., Morabito, F. C., Kasabov, N. (2016, July). Longitudinal study of alzheimer's disease degeneration through EEG data analysis with a NeuCube spiking neural network model. In Neural Networks (IJCNN), 2016 International Joint Conference on (pp. 1360-1366). IEEE.
- Kasabov, N., Scott, N. M., Tu, E., Marks, S., Sengupta, N., Capecci, E., Othman, M., Gholoami Doborjeh, M., Murli, N., Hartono, R., Espinosa-Ramos, J. I., Zhou, L., Alvi, F., Wang, G., Taylor, D., Feigin, V., Gulyaev, S., Mahmoud, M., Hou, Z. G., Yang, J. (2016). Evolving spatio-temporal data machines based on the NeuCube neuromorphic framework: design methodology and selected applications. Neural Networks, 78, 1-14.
R&D system
For this project, an R&D system has been developed based on NeuCube. The system can be obtained subject to licensing agreement.
Developers
A new, generic methodology for mapping, learning, visualisation, classification and understanding of fMRI data using the NeuCube architecture of SNN. A solution to a problem defined by fMRI data is not a single formula or an algorithm, but an evolving spatio-temporal data machine (eSTDM) that consists of several modules, each of them having a set of alternative algorithms and parameter values that can be optimised.
The Brain-Like Artificial Intelligence (BLAI) is pioneered by Professor Nikola Kasabov and here it is applied to a specific application.
The project introduces a new methodology and a system for dynamic learning, visualisation and classification of functional Magnetic Resonance Imaging (fMRI) as Spatio-Temporal Brain Data (STBD). The method is based on an evolving spatio-temporal data machine (eSTDM) of evolving spiking neural networks (SNN) exemplified by the NeuCube architecture. The method consists of several steps: mapping spatial coordinates of fMRI data into a 3D SNN cube that represents a brain template; input data transformation into trains of spikes; deep, unsupervised learning in the 3D SNN cube of spatio-temporal patterns from data; supervised learning in an evolving SNN classifier; parameter optimisation; 3D visualisation and model interpretation. The learned connections in the SNN cube represent dynamic spatio-temporal relationships derived from the fMRI data. They can reveal new information about the underpinning brain functions under different conditions. The proposed methodology allows for the first time to analyse dynamic functional and structural connectivity of a learned SNN model from fMRI data. This can be used for a better understanding of brain activities and also for on-line generation of appropriate neuro-feedback to subjects for improved brain functions. The proposed NeuCube-based methodology offers also a superior classification accuracy when compared with traditional AI and statistical methods. The created NeuCube-based models of fMRI data are directly and efficiently implementable on high performance and low energy consumption neuromorphic platforms for real–time applications.
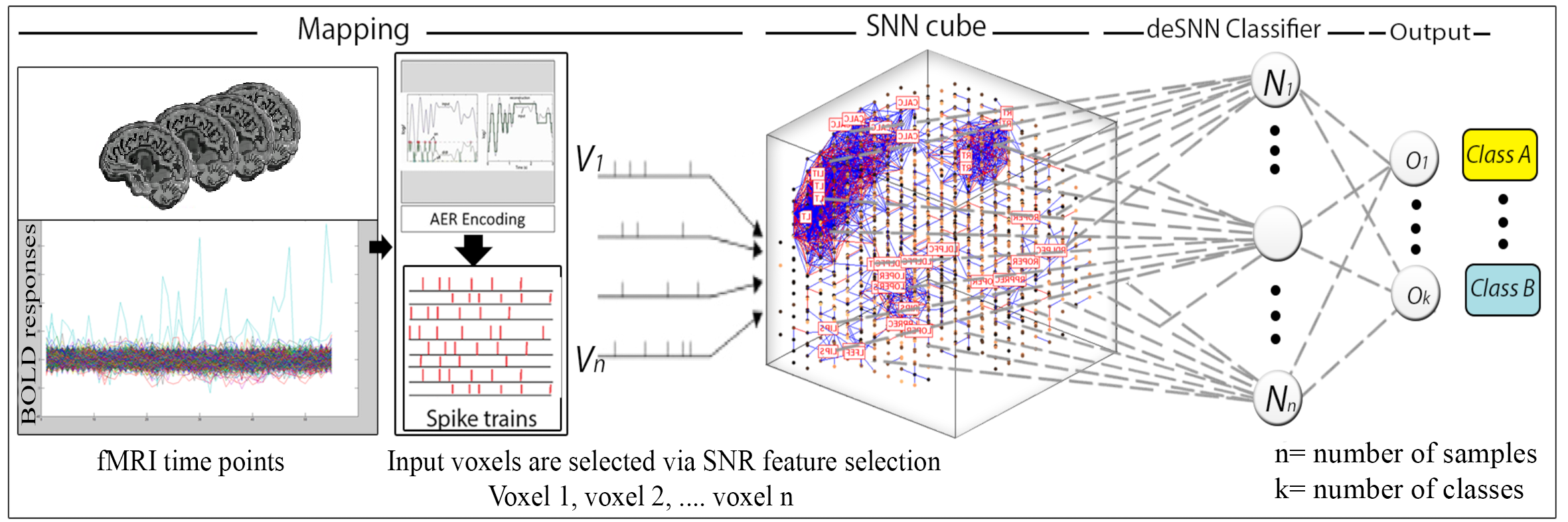
A block diagram of the NeuCube SNN model for fMRI data encoding, mapping, learning, classification and visualisation.
Related papers and benchmarking
The proposed methods and systems, when compared with traditional statistical and machine learning methods, showed superior results in the following aspects:
- An efficient neuromorphic approach, which offers better interpretation of the created models from one of the most complex STBD—fMRI data;
- Better classification accuracy when compared with traditional AI and statistical methods (see table below from Kasabov, Doborjeh, & Doborjeh, 2017);
- Reveals trajectories of functional spatio-temporal associations between areas of the brain (none of the traditional methods can reveal such interactions);
- Better visualisation of the created models, with a possible use of VR;
- Enabling new information and knowledge discovery through meaningful interpretation of the models.
Measure | SVM | MLP | ECF | ECMC | MLR | NeuCube |
|---|---|---|---|---|---|---|
Accuracy (%) | 85 | 76 | 78 | 77 | 68 | 87 |
F -Score | 0.51 | 0.46 | 0.57 | 0.59 | 0.57 | 0.93 |
See also some of the related papers:
- Kasabov, N. K., Doborjeh, M. G., & Doborjeh, Z. G. (2017). Mapping, learning, visualization, classification, and understanding of fMRI Data in the NeuCube evolving spatiotemporal data machine of spiking neural networks. IEEE transactions on neural networks and learning systems, 28(4), 887-899.
- Kasabov, N., Zhou, L., Doborjeh, M. G., Doborjeh, Z. G., & Yang, J. (2016). New Algorithms for encoding, learning and classification of fmri data in a spiking neural network architecture: a case on modelling and understanding of dynamic cognitive processes.IEEE Transactions on Cognitive and Developmental Systems.
fMRI data
Data can be download here.
R&D system
For this project, an R&D system has been developed based on NeuCube. The system can be obtained subject to licensing agreement.
Developer
This project develops novel methods and systems for BCI.
The Brain-Like Artificial Intelligence (BLAI) is pioneered by Professor Nikola Kasabov and here it is applied to a specific application.
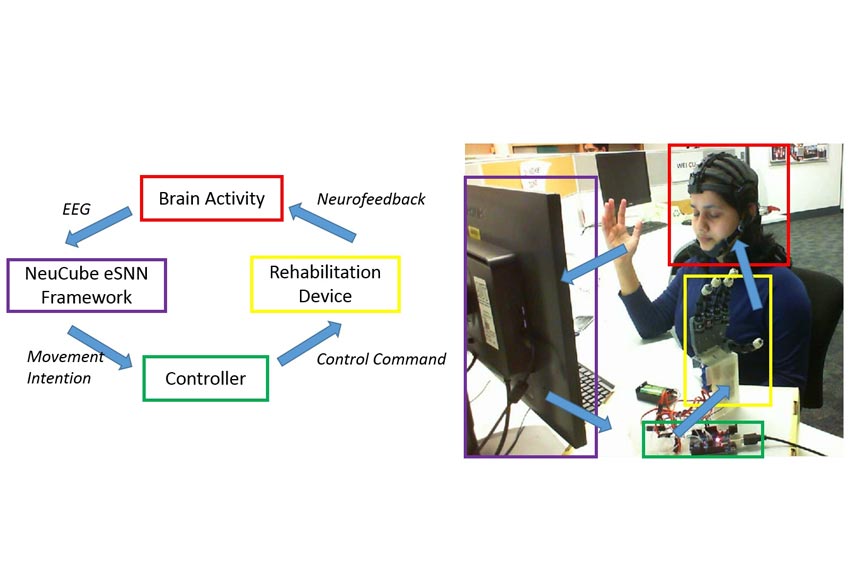
Brain computer interface (BCI) monitors the neural activities of the brain and translate them to machine commands to control devices such as computers, wheelchairs or robots. Recent studies have shown the feasibility of BCI to assist the activities of daily living in paralyzed patients. NeuCube’s advanced data analytics capabilities on analysing spatio-temporal brain data have shown promising empirical results on decoding motor tasks from electroencephalography (EEG) signals. This study shows the feasibility of NeuCube spiking neural network architecture for developing a functional brain computer interface platform to enable robot assisted neurorehabilitation through BCI. Through this approach we aim to detect the patient’s intention to move his or her hand and pass the command to control an exoskeleton or functional eectric stimulation (FES) system.
Decoding movements of the same limb is an important problem in BCI for Neurorehabilitation. Due to the non‐invasiveness and high temporal resolution, EEG has been widely used for decoding movements in BCI. However, less spatial resolution caused by the limited number of electrodes is a challenge for pattern recognition. Previous studies on neural activities in motor related areas of the brain during physical movements provide evidences that approximately the same area of brain is activated during the movements of the same limb. Thus, classification of movements of same limb from EEG results less accuracy and limits applicability of BCI for Neurorehabilitation. The state-based online classification module of NeuCube addresses this limitation and facilitates a BCI platform for Neurorehabilitation. The module facilitates a brain state based classification of EEG signals using spiking neural networks (SNN). The module encloses a finite state machine (FSM) which acts as a finite memory to the model and a biologically plausible NeuCube SNN architecture to decode state transitions over the time. We performed offline training of NeuCube model using pre-recorded EEG data through spike time dependant plasticity (STDP) learning rule. The trained NeuCube model is then used to perform online classification on EEG data streams using the dynamic evolving spiking neural network (deSNN) classifier. The module follows the cue based (synchronous) BCI paradigm. The subject is required to perform a motor task (i.e. hand opening, hand closing) according to the cue displayed on the screen. While the subject is performing the task, EEG signals are recorded and classified. This classification output is used to control the rehabilitation robot arm. This approach enables the user to control the rehabilitation robots through their own thoughts and intentions and provides neurofeedback to help patients to improve their brain functions.
Development of spiking neural network based computational models and algorithms for brain computer interfaces
Two computational models that use spiking neural networks were proposed to address the limitations in current approaches for BCI control. These models are:
- FaNeuRobot: We propose a ‘brain-inspired’ framework for continuous motor control through Brain Computer Interface using Finite Automata theory and NeuCube evolving SNN architecture. This model addresses the less spatial resolution of EEG particularly evident during different movements of the same upper limb.
- eSPANet: Evolving spike pattern association networks for spike-based supervised incremental learning
Due to the non-stationarity and high trial-to-trial variability of brain data, online classification is challenging for BCI. This is more significant when BCI is applied to neurological rehabilitation where the person incrementally learns to regain the control of movement. Here we have proposed a computational model that mimics the incremental learning in biological neural networks using a network of spiking neurons. The model contains a group of spike pattern association neurons, a spiking neuron model which is able to emit spikes at the desired time-point. According to the stimulus, these neurons are assigned to the relevant neuron population referred as a ‘population vector’. The readout neuron will be trained to produce the corresponding response for the incoming input signals.
Related papers and benchmarking
The proposed methods and systems, when compared with traditional statistical and machine learning methods, showed superior results in the following aspects:
- Better data analysis and classification accuracy compared to state of the art machine learning methods such as Support Vector Machine, Multi-Layer Perceptron and Linear Discriminant Analysis (Table1).
- Better visualisation of the created models through the dynamic visualisation module of NeuCube and as well as with a possible use of VR;
- Better understanding of the data and the processes that are measured;
- Enabling new information and knowledge discovery through meaningful interpretation of the models.
Measure | SVM | MLP | LDA | NeuCube |
|---|---|---|---|---|
Accuracy (%) - Session 1 | 55% | 85% | 65% | 90% |
Accuracy (%) - Session 2 | 45% | 75% | 85% | 95% |
Average Accuracy (%) | 50% (+-7%) | 80% (+-7%) | 75% (+-14%) | 92.5% (+-3.5%) |
See also some of the related papers:
- Kasabov, N. K. (2014). NeuCube: A spiking neural network architecture for mapping, learning and understanding of spatio-temporal brain data. Neural Networks, 52, 62-76.
- Kasabov, N., Scott, N. M., Tu, E., Marks, S., Sengupta, N., Capecci, E., Othman, M., Gholoami Doborjeh, M., Murli, N., Hartono, R., Espinosa-Ramos, J. I., Zhou, L., Alvi, F., Wang, G., Taylor, D., Feigin, V., Gulyaev, S., Mahmoud, M., Hou, Z. G., Yang, J. (2016). Evolving spatio-temporal data machines based on the NeuCube neuromorphic framework: design methodology and selected applications. Neural Networks, 78, 1-14.
- Hu, J., Hou, Z. G., Chen, Y. X., Kasabov, N., & Scott, N. (2014, August). EEG-based classification of upper-limb ADL using SNN for active robotic rehabilitation. In Biomedical Robotics and Biomechatronics (2014 5th IEEE RAS & EMBS International Conference on (pp. 409-414). IEEE.
- Taylor, D., Scott, N., Kasabov, N., Capecci, E., Tu, E., Saywell, N.,Chen, Y., Hu, J., Hou, Z. G. (2014, July). Feasibility of neucube snn architecture for detecting motor execution and motor intention for use in bci applications. In Neural Networks (IJCNN), 2014 International Joint Conference on (pp. 3221-3225). IEEE.
- Kumarasinghe, k., Owen, M., Taylor, D., Kasabov, N., and Au, C.K. “FaNeuRobot: A ‘Brain-like’ Framework for Robot and Prosthetics Control using the NeuCube Spiking Neural Network Architecture & Finite Automata Theory,” IEEE International Conference on Robotics and Automation 2018. (Submitted).
- Kumarasinghe, K., Taylor, D., and Kasabov, N. “eSPANNet: Evolving Spike Pattern Association Neural Network for Spike-based Supervised and Incremental Learning and Its Application on Single-trial Brain Computer Interfaces,” IEEE Transactions on Neural Networks and Learning Systems. (Submitted).
R&D system
For this project, an R&D system has been developed based on NeuCube. The system can be obtained subject to licensing agreement.
Our research groups
The research and development work done by KEDRI's founding director, Professor Nikola Kasabov, and his team is organised into six areas of research.